It is a common practice in the statistical analysis of data to group the samples into bins or categories. The goal is usually to extract some structure that is useful and often hidden from us when looking at direct plots of the sample set.
For instance, a time series plot of the change in stock price may appear erratic and contain no order, but observing the frequency of certain price-changes may help us realize that on average there is no change as well as give us an idea as to the practical bounds to how much the price will change.
The purpose of this note is to speak a little to the role of histograms in statistics and probability. While a variety of complications arise when using them for the estimation of a probability density function, they serve as a very practical (and efficient) tool in exploratory data analysis and provide a grounding for other forms of density estimation.
Histogram Construction
- For a given a sample, what bin does it belong to?
- How many samples are in a given bin?
- What is the numerical range or volume of a given bin?
From here forward, we consider data samples which can lie in some region of the real number line (e.g. integers, fractions, and roots of integers). We’ll represent a sample of numbers by the vector  , say
, say 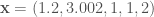 .
.
 The diagram outlines some relevant quantities of a histogram. The minimum and maximum values of the sample (1 and 3.002 in the example above) help define the range that we will distribute our bins in. We represent the bin width as
The diagram outlines some relevant quantities of a histogram. The minimum and maximum values of the sample (1 and 3.002 in the example above) help define the range that we will distribute our bins in. We represent the bin width as 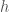 . It is not required that a histogram have fixed-width bins, but this is one of the most commonly used and useful formulations.
. It is not required that a histogram have fixed-width bins, but this is one of the most commonly used and useful formulations.
The starting offset (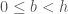 ) is also a useful quantity. For instance, if the first bin starts at
) is also a useful quantity. For instance, if the first bin starts at  or
or 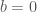 , any number falling in the range (
, any number falling in the range ( ) contributes to the quantity of elements in that bin. For
) contributes to the quantity of elements in that bin. For  , the range of the first bin in the example above is (
, the range of the first bin in the example above is (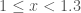 ). The two 1s and 1.2 will fall in the first bin.
). The two 1s and 1.2 will fall in the first bin.
Suppose we shift the bins a little to the left (i.e. 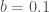 ). The range of the first bin is now (
). The range of the first bin is now ( ). The two 1s fall in the first bin while 1.2 falls in the second. So it is easy to see how both the bin width and offset affect our view of the histogram.
). The two 1s fall in the first bin while 1.2 falls in the second. So it is easy to see how both the bin width and offset affect our view of the histogram.
The total number of bins in a histogram can be represented as:
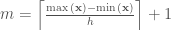
where the  represents the next integer larger than
represents the next integer larger than  , and this follows analogously for
, and this follows analogously for 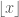 . We add 1 to the traditional bin count to accommodate our offset
. We add 1 to the traditional bin count to accommodate our offset 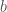 .
.
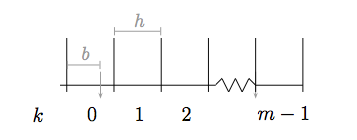
When constructing a histogram on a computer, we desire a function 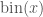 that maps a sample to an appropriate bin index
that maps a sample to an appropriate bin index  . Assuming that we begin the bins at 0, the bin index can be determined by dividing the sample from the bin width:
. Assuming that we begin the bins at 0, the bin index can be determined by dividing the sample from the bin width: 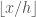 . For
. For  , it’s easy to see that numbers less than 0.3 will fall in bin 0, while a number twice as large will fall in the second bin, and so on. Accounting for the offset, we have:
, it’s easy to see that numbers less than 0.3 will fall in bin 0, while a number twice as large will fall in the second bin, and so on. Accounting for the offset, we have:

We also desire a function that will allow us to compute the start of the interval for a given bin index:

The formulation above allows us to experiment with the two essential qualities of a histogram (bin width and offset) and it should be straightforward to see how this simple approach generalizes for multi-dimensional histograms of equal bin widths, except we then consider more generally the bin volume (e.g in the three-dimensional case the volume  ).
).
In the terminology of computer science, these are constant time procedures. If the bin widths are not fixed and arbitrarily set by the user then a more complicated method is required. One can use a specialized function or binary search for finding the appropriate bin in this case [1]. Histograms can also accept a variety of types (e.g. integers, single- or double-precision floating-point numbers) as inputs and if the compiler knows about this information ahead of time, it can optimize its code even further while providing a general interface of accumulators [2].
Histograms as density estimates
We now turn to a more mathematical perspective on histograms. It is natural to treat a histogram as the estimation of a probability density function. Once armed with the density function a variety of tools from probability and information theory are at our disposal, but we must take caution that the estimates of the density function hold true to the data sample and question whether we should be estimating the densities in the first place to solve the desired problem.
To get a taste for what this is like, we will derive the maximum likelihood estimator for a mathematical step function that represents the histogram.

Here, the function has  parameters:
parameters: 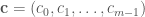 . Provided our data sample and a given bin
. Provided our data sample and a given bin  , what is the optimal height,
, what is the optimal height,  , of the step function?
, of the step function?
To find an estimate of the density function 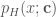 we express the likelihood of our sample of data given the model parameters as:
we express the likelihood of our sample of data given the model parameters as:

Here,  represents the number of observed samples in bin
represents the number of observed samples in bin  . In other words, we’d like find
. In other words, we’d like find  that maximizes the probability that we’ll find
that maximizes the probability that we’ll find  items in bin
items in bin  . This optimization is subject to the density function constraint:
. This optimization is subject to the density function constraint:
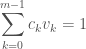
where  is the volume of the given histogram region.
is the volume of the given histogram region.
Constrained optimizations of this form can be solved using the method of Lagrange multipliers. In this case, we have a multidimensional function:

subject to the constraint that:

Note that we took the log of the likelihood function so we can deal with sums rather than products. This will come in handy when taking derivatives in a moment and is a common practice.
The gradient of a multidimensional function results in a vector field where each point–in our case represented by  –has an associated vector of steepest ascent or descent on the multidimensional surface [3]. Our goal is to find where the steepest ascent of
–has an associated vector of steepest ascent or descent on the multidimensional surface [3]. Our goal is to find where the steepest ascent of  is equal in magnitude and direction to that of
is equal in magnitude and direction to that of  . This equality is represented as:
. This equality is represented as:

And we can express this combined optimization with:

where  is called the “Lagrangian”. Computing the derivatives:
is called the “Lagrangian”. Computing the derivatives:

From this, we can see that

Since  and
and  ,
,  . Thus our optimal height and estimate of the density is:
. Thus our optimal height and estimate of the density is:

While this estimator is actually quite an intuitive result, it is important to observe that this only in the special case of step functions with varying volumes predefined. What if, for example, we wanted to see what the best estimator is given a piecewise linear function or a spline between bins? Or what if we want to find out what the bin volumes  should be as well? This question motivates a general discussion on the estimation of the probability density function given the data sample
should be as well? This question motivates a general discussion on the estimation of the probability density function given the data sample  , and these “data-driven” approaches are dubbed non-parametric [4].
, and these “data-driven” approaches are dubbed non-parametric [4].
Similar issues as those brought up in density estimation also arise in the estimation of mutual information [5].
—–
[1] Given a sample, start at the center-most bin. If the sample is greater than that value, then start at the center most bin of the right half, otherwise do the same to the left. Recurse this procedure until the desired bin is found. The running time of this scales like  . The GNU scientific library provides utilities in this spirit
. The GNU scientific library provides utilities in this spirit
[2] See the C++ Boost Accumulator library for more information on this.
[3] For a more detailed tour of why this is as well as an intuitive contour plot geometrical interpretation of this, consult an introductory applied math text. Setting 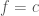 , we can study the function’s “surface contours”. By definition, a vector
, we can study the function’s “surface contours”. By definition, a vector  along this contour does not change the value of
along this contour does not change the value of  , therefore
, therefore  . Because of this,
. Because of this,  must be normal to
must be normal to  .
.
[4] Scott DW, Sain SR. “Multidimensional density estimation”. Handbook of Statistics: Data Mining and Computational Statistics 23. 297-306 (2004). Gentle JE. “Nonparametric estimation of probability density functions”. Elements of Computational Statistics. 205-231 (2002).
[5] Paninski L. “Estimation of entropy and mutual information”. Neural Computation 15. 1191-1253 (2003). Fraser AM, Swinney HL. “Independent coordinates for strange attractors from mutual information”. Physical Review A 33. 1334-1140 (1986).




